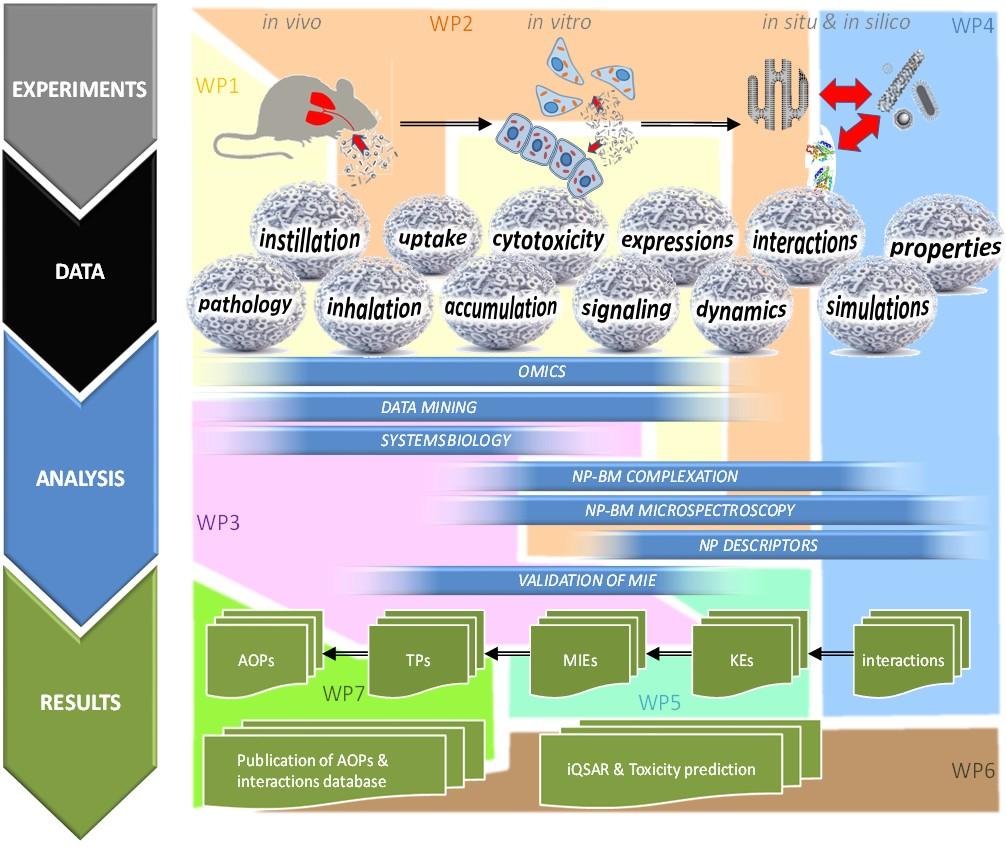
SmartNanoTox workpackages
Our research aims are based around the following objectivies:
- To identify main pulmonary adverse outcomes induced by common NMs, and identify associated MIE, KEs and toxicity pathways leading to AO.
- To establish relationships between physicochemical properties of NMs and KEs steering the TP leading to AO, and suggest descriptors for grouping of NMs according to their toxicological mode-of-action.
- To create a database of bionano interactions that will enable development of read-across and QSAR tools for the toxicity assessment of new NMs.
- To develop a smart screening approach, where predictions of toxicity of a NM can be made on the basis of purely computational or limited in vitro screening tests focused on crucial bionano interactions.
The research programme combines a broad range of experimental and theoretical methods covering all stages of NM interaction with an organism from the initial uptake to the (post contact) adverse outcome. We attempt to identify all possible short-term and long-term adverse effects related to inhalation for a range of selected NMs. We focus on a relatively small number of materials and exposure routes to be able to achieve the mechanistic interpretation of the NM transport, crucial bionano interactions that can trigger the adverse outcome pathway (AOP), and thus relate the basic NM descriptors to the adverse effects. We expect that the results of this research will enable the subsequent formulation of intelligent QSAR systems based on key nanoparticle descriptors. Based on the analysis of the NM-induced pathways, the project will develop toxicity pathway-based QSARs and map the NM physicochemical properties to the MIE/KE and thus to the specific AO for any NM.
- Identification and classification of the entry mechanisms and adverse effects of NMs in vivo and in vitro.
This WP deals with in vivo exposure supplemented with in vitro experiments: inhalation, instillation of rodents, analysis of the gene expression and proteomics and supplies the data about the NM entry routes and endpoints for WP2 and WP3.
- Identification and classification of the state of nanoparticles after initial contact with the organism.
This WP contains in vitro experiments, either complementing the in vivo studies of WP1 or standalone and aims at characterising the NM state after entering the organism, or specific molecular mechanisms of the uptake, transport or toxicity
- Identification of end-points and classification of the possible TPs, identification of the MIEs and KEs for selected pathways.
This WP is devoted to analysis and integration of the gene expression and/or proteomics data as obtained in WP1 and WP2, data mining and statistical modelling aiming to establish the adverse outcome pathways and identify the MIE/KEs
- Database and QSAR for NM-biomolecule interactions.
This WP is computational and deals with modelling biomolecules in contact with nanoparticles and building a database of bionano interactions.
- Assessment and validation of the AOPs and MIE/KE, test of NM descriptors as predictors.
WP5 assesses the candidate MIE/KEs as extracted in WP3 either in silico, using the models from WP4, or in vitro and validates the identified pathways and MIE/KE by modifying either the NM or the target system
- Development of a smart test addressing the molecular key events.
Here we finally construct a simplified in vitro or in silico test for the identified AOPs using the database from WP4 combined with findings of WP5.
- Dissemination and exploitation.
- WP8 Management.
This diagram below shows the flow fo the work packages mentioned above